Recent studies have found that
Gires-Tournois (GT) biosensors, a type of nanophotonic resonator, can detect
minuscule virus particles and produce colorful micrographs (images taken
through a microscope) of viral loads. But they suffer from visual artifacts and
non-reproducibility, limiting their utilization.
In a recent breakthrough, an international team of researchers, led by Professor Young Min Song from the School of Electrical Engineering and Computer Science at Gwangju Institute of Science and Technology in Korea, has leveraged artificial intelligence (AI) to overcome this problem. Their work was published in Nano Today.
Rapid and on-site diagnostic technologies for identifying and quantifying viruses are essential for planning treatment strategies for infected patients and preventing further spread of the infection. The COVID-19 pandemic has highlighted the need for accurate yet decentralized diagnostic tests that do not involve complex and time-consuming processes needed for conventional laboratory-based tests.
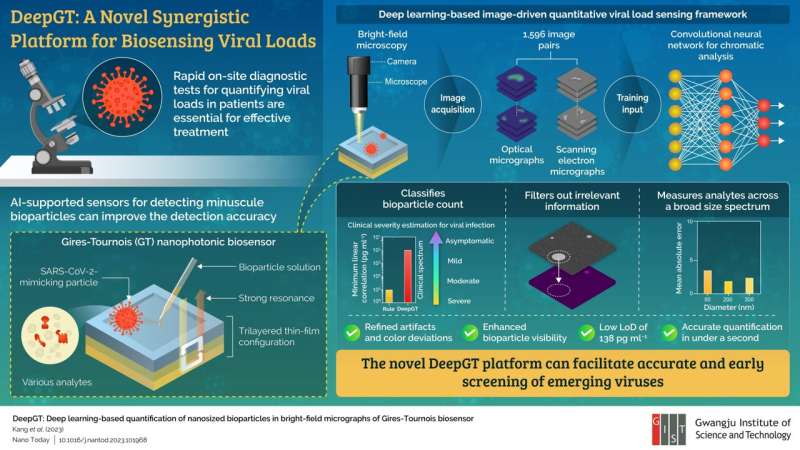
A popular point-of-care diagnostic tool for quantifying viral loads is bright-field microscopic imaging. However, the small size (~ 100 nm) and low refractive index (~ 1.5, same as that of a microscope slide) of bioparticles such as viruses often makes their accurate estimation difficult and increases the limit of detection (the lowest concentration of viral load that can be reliably detected).
In their new study, the team proposed a synergistic biosensing tool called "DeepGT," which can harness the advantages of GT sensing platforms and merge them with deep learning-based algorithms to accurately quantify nanoscale bioparticles, including viruses, without the need for complex sample preparation methods.
"We designed DeepGT to objectively assess the severity of an infection or disease. This means that we will no longer have to rely solely on subjective assessments for diagnosis and health care but will instead have a more accurate and data-driven approach to guide therapeutic strategies," explains Prof. Song, revealing the motivation behind their study.
The team designed a GT biosensor with a trilayered thin-film configuration and biofunctionalized it to enable colorimetric sensing upon interaction with target analytes. The sensing abilities were verified by simulating the binding mechanism between host cells and the virus using specially prepared bioparticles that mimicked SARS-CoV-2—the coronavirus strain that caused the COVID-19 pandemic.
Next, the researchers trained a convolutional neural network (CNN) using over a thousand optical and scanning electron micrographs of the GT biosensor surface with different types of nanoparticles. They found that DeepGT was able to refine visual artifacts associated with bright-field microscopy and extract relevant information, even at viral concentrations as low as 138 pg ml–1.
Moreover, it determined the bioparticle count with a high accuracy, characterized by a mean absolute error of 2.37 across 1,596 images compared to 13.47 for rule-based algorithms, in under a second. Boosted by the performance of CNNs, the biosensing system can also indicate the severity of the infection from asymptomatic to severe based on the viral load.
DeepGT thus presents an efficient and precise way of screening viruses across a broad size range without being hindered by the minimum diffraction limit in visible light. "Our approach provides a practical solution for the swift detection and management of emerging viral threats as well as the improvement of public health preparedness by potentially reducing the overall burden of costs associated with diagnostics," concludes Prof. Song.

 Previous page
Previous page Back to top
Back to top







