Most organisms use oxygen to
convert food into energy. However, in environments with little or no oxygen,
life found other ways to produce energy, using a process called fermentation.
Fermentative metabolism occurs in environments such as ruminant guts, sediments, and anaerobic bioreactors. Aside from producing energy that the organisms can use to thrive, the process can result in other end products including biofuels and other bioproducts.
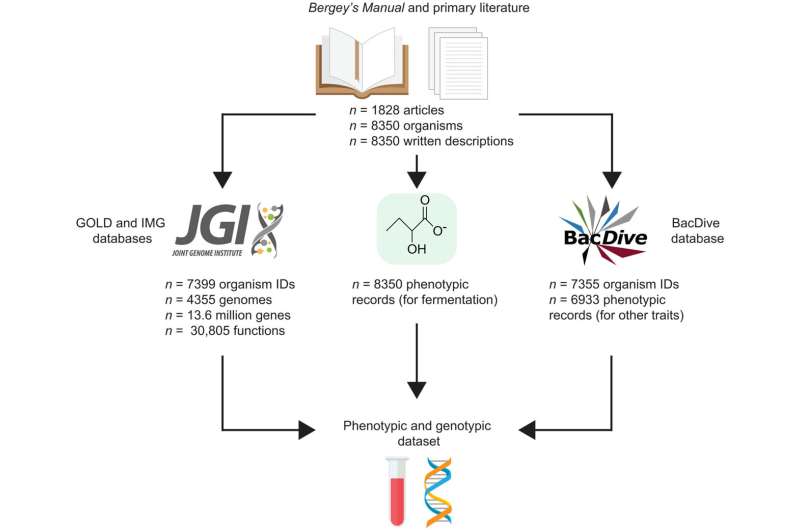
To better understand the range of bacteria and archaea that rely on this form of metabolism, researchers recently compiled a list of with more than 8,300 organisms from multiple sources, including the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility located at Lawrence Berkeley National Laboratory (Berkeley Lab), and the DOE Systems Biology Knowledgebase (KBase).
They also built an interactive browser that allows other researchers to study the genomes and predict the metabolic abilities of microbes that are likely to thrive in environments with little or no oxygen. The study is published in Science Advances.
Researchers at the University of California, Davis and UC Santa Barbara generated a list of prokaryotes with fermentative metabolism from the literature. They then used the JGI's GOLD and IMG data portals to get information about the genomes and genes of the organisms on the list. They also searched IMG/M for all protein-coding genes belonging to each genome.
The final dataset—available to the research community on an online interactive browser—contains the phenotypic records of 8,350 organisms along with 4,355 genomes and 13.6 million genes. The results reveal that fermentative metabolism is seen in a third of prokaryotes and can form roughly 300 combinations of metabolites.
To demonstrate the potential utility of the browser, the team used Fermentation Explorer with a series of large datasets. One test was to predict traits of more than 400 cultured rumen prokaryotes from the Hungate1000 project. They also used the tool to predict possible functions from 733 metagenome-assembled rumen genomes. Finally, they used the tool to predict the fermentative end products of five previously uncharacterized prokaryotes, including two bacteria isolated by their lab.
KBase was used to rapidly assemble and annotate the taxonomy and function of these two isolates. The combined taxonomic and functional annotations were used to predict the final end products from these genomes. The team then cultured the organisms and found general agreement between the predicted and actual end products.

 Previous page
Previous page Back to top
Back to top







